|
SNAP Library 2.1, Developer Reference
2013-09-25 10:47:25
SNAP, a general purpose, high performance system for analysis and manipulation of large networks
|
|
SNAP Library 2.1, Developer Reference
2013-09-25 10:47:25
SNAP, a general purpose, high performance system for analysis and manipulation of large networks
|
#include <agmfit.h>
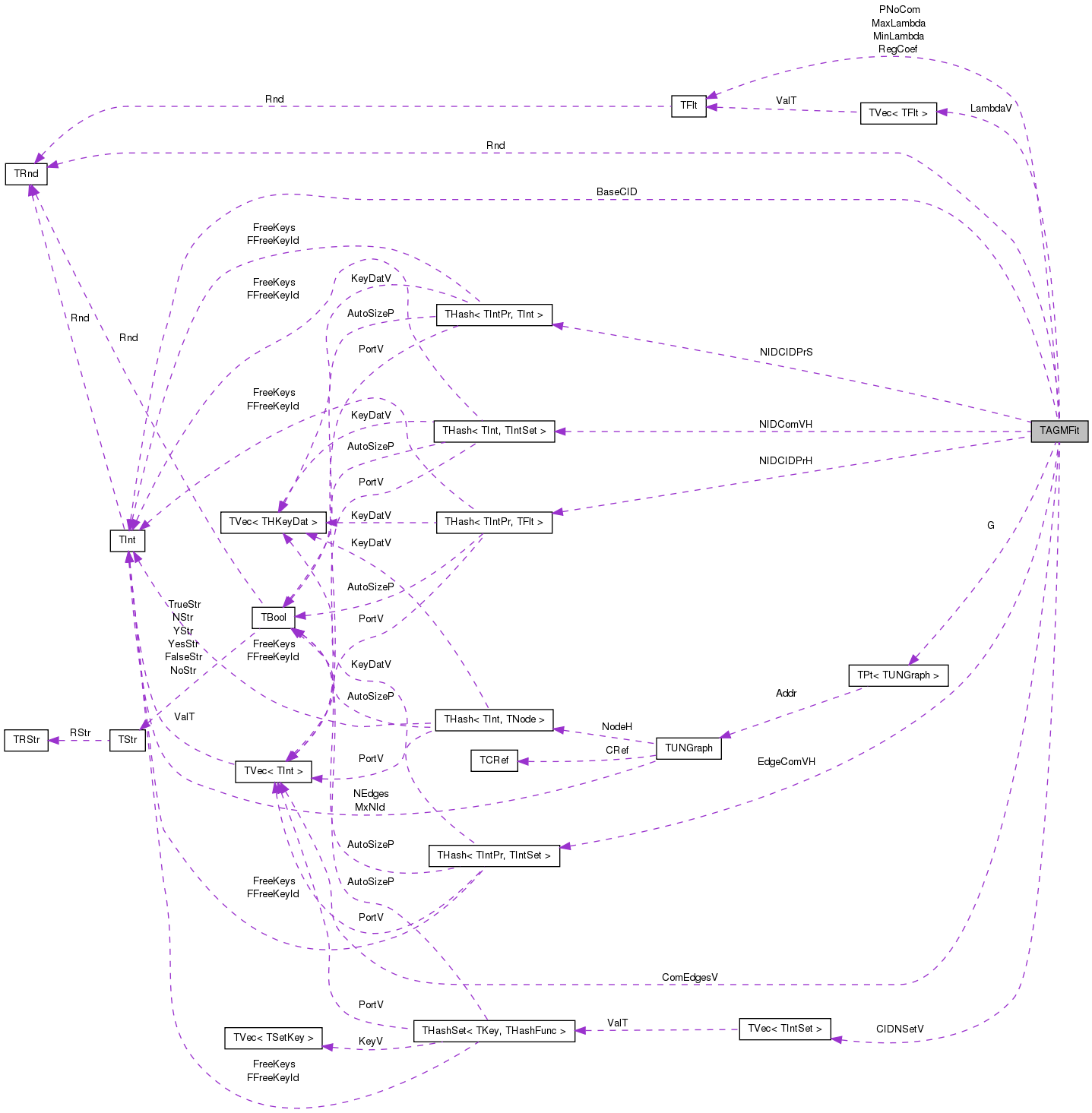
Public Member Functions | |
| TAGMFit () | |
| ~TAGMFit () | |
| TAGMFit (const PUNGraph &GraphPt, const TVec< TIntV > &CmtyVVPt, const int RndSeed=0) | |
| COMMENT. Use to describribe parameters. | |
| TAGMFit (const PUNGraph &GraphPt, const int InitComs, const int RndSeed=0) | |
| COMMENT. Use to describribe parameters. | |
| TAGMFit (const PUNGraph &GraphPt, const TVec< TIntV > &CmtyVVPt, const TRnd &RndPt) | |
| COMMENT. Use to describribe parameters. | |
| void | Save (TSOut &SOut) |
| void | Load (TSIn &SIn, const int &RndSeed=0) |
| void | RandomInitCmtyVV (const int InitComs, const double ComSzAlpha=1.3, const double MemAlpha=1.8, const int MinComSz=8, const int MaxComSz=200, const int MinMem=1, const int MaxMem=10) |
| Randomly initialize bipartite community affiliation graph. | |
| void | AddBaseCmty () |
| Add Epsilon community (base community which includes all nodes) into community affiliation graph. This means that we will later fit the value of epsilon. | |
| double | Likelihood () |
| COMMENT. | |
| double | Likelihood (const TFltV &NewLambdaV) |
| double | Likelihood (const TFltV &NewLambdaV, double &LEdges, double &LNoEdges) |
| void | SetRegCoef (const double Val) |
| void | GetEdgeJointCom () |
For each (u, v) in edges, precompute C_uv (the set of communities nodes u and v share). | |
| void | NeighborComInit (const int InitComs) |
| Initialize node community memberships using best neighborhood communities (see D. Gleich et al. KDD'12). | |
| void | GradLogLForLambda (TFltV &GradV) |
| int | MLEGradAscentGivenCAG (const double &Thres=0.001, const int &MaxIter=10000, const TStr PlotNm=TStr()) |
Gradient descent for p_c while keeping the community affiliation graph (CAG) fixed. | |
| void | SetDefaultPNoCom () |
| Set Epsilon (the probability that two nodes sharing no communities connect) to the default value. | |
| void | SetPNoCom (const double &Epsilon) |
| Set Epsilon (the probability that two nodes sharing no communities connect) to the default value. | |
| double | GetPNoCom () |
| double | CalcPNoComByCmtyVV (const int &SamplePairs=-1) |
| Compute the empirical edge probability between a pair of nodes who share no community (epsilon), based on current community affiliations. | |
| void | GetNewtonStep (TFltVV &HVV, TFltV &GradV, TFltV &DeltaLV) |
| double | SelectLambdaSum (const TIntSet &ComK) |
Compute sum of lambda_c (which is log (1 - p_c)) over C_uv (ComK). The function is used to compute edge probability P_uv. | |
| double | SelectLambdaSum (const TFltV &NewLambdaV, const TIntSet &ComK) |
| COMMENT. | |
| void | RandomInit (const int &MaxK) |
| COMMENT. | |
| void | RunMCMC (const int &MaxIter, const int &EvalLambdaIter, const TStr &PlotFPrx=TStr()) |
| Main procedure for fitting the AGM to a given graph using MCMC. | |
| void | SampleTransition (int &NID, int &JoinCID, int &LeaveCID, double &DeltaL) |
Sample MMCM transitions: Choose among (join, leave, switch), and then sample (NID, CID). | |
| void | InitNodeData () |
| COMMENT. | |
| void | LeaveCom (const int &NID, const int &CID) |
After MCMC, NID leaves community CID. | |
| void | JoinCom (const int &NID, const int &JoinCID) |
| int | RemoveEmptyCom () |
| Remove all communities with no members. | |
| double | SeekLeave (const int &UID, const int &CID) |
Compute the change in likelihood (Delta) if node UID leaves community CID. | |
| double | SeekJoin (const int &UID, const int &CID) |
Compute the change in likelihood (Delta) if node UID joins community CID. | |
| double | SeekSwitch (const int &UID, const int &CurCID, const int &NewCID) |
| double | GetStepSizeByLineSearchForLambda (const TFltV &DeltaV, const TFltV &GradV, const double &Alpha, const double &Beta) |
| Step size search for updating P_c (which is parametarized by regularization parameter lambda). | |
| void | SetLambdaV (const TFltV &LambdaPt) |
| COMMENT. | |
| void | GetLambdaV (TFltV &OutV) |
| COMMENT. | |
| void | GetQV (TFltV &OutV) |
Returns QV, a vector of (1 - p_c) for each community c. | |
| void | GetCmtyVV (TVec< TIntV > &CmtyVV, const double QMax=2.0) |
Get communities whose p_c is higher than 1 - QMax. | |
| void | GetCmtyVV (TVec< TIntV > &CmtyVV, TFltV &QV, const double QMax=2.0) |
| COMMENT. | |
| void | SetCmtyVV (const TVec< TIntV > &CmtyVV) |
| COMMENT. | |
| void | PrintSummary () |
| COMMENT. | |
Private Attributes | |
| PUNGraph | G |
| Graph to fit. | |
| TVec< TIntSet > | CIDNSetV |
| Community ID -> Member Node ID Sets. | |
| THash< TIntPr, TIntSet > | EdgeComVH |
| Edge -> Shared Community ID Set. | |
| THash< TInt, TIntSet > | NIDComVH |
| Node ID -> Communitie IDs the node belongs to. | |
| TIntV | ComEdgesV |
| The number of edges in each community. | |
| TFlt | PNoCom |
| Probability of edge when two nodes share no community (epsilon in the paper). | |
| TFltV | LambdaV |
| Parametrization of P_c (edge probability in community c), P_c = 1 - exp(-lambda). | |
| TRnd | Rnd |
| THash< TIntPr, TFlt > | NIDCIDPrH |
| <Node ID, Community ID> pairs (for sampling MCMC moves). | |
| THash< TIntPr, TInt > | NIDCIDPrS |
| <Node ID, Community ID> pairs (for sampling MCMC moves). | |
| TFlt | MinLambda |
| Minimum value of regularization parameter lambda (default = 1e-5). | |
| TFlt | MaxLambda |
| Maximum value of regularization parameter lambda (default = 10). | |
| TFlt | RegCoef |
| Regularization parameter when we fit for P_c (for finding # communities). | |
| TInt | BaseCID |
| ID of the Epsilon-community (in case we fit P_c of the epsilon community). We do not fit for the Epsilon-community in general. | |
| TAGMFit::TAGMFit | ( | ) | [inline] |
| TAGMFit::~TAGMFit | ( | ) | [inline] |
| TAGMFit::TAGMFit | ( | const PUNGraph & | GraphPt, |
| const TVec< TIntV > & | CmtyVVPt, | ||
| const int | RndSeed = 0 |
||
| ) | [inline] |
| TAGMFit::TAGMFit | ( | const PUNGraph & | GraphPt, |
| const int | InitComs, | ||
| const int | RndSeed = 0 |
||
| ) | [inline] |
COMMENT. Use to describribe parameters.
Definition at line 31 of file agmfit.h.
References NeighborComInit().
: G(GraphPt), PNoCom(0.0), Rnd(RndSeed), MinLambda(0.00001), MaxLambda(10.0), RegCoef(0), BaseCID(-1) { NeighborComInit(InitComs); }//RandomInitCmtyVV(InitComs); }
| TAGMFit::TAGMFit | ( | const PUNGraph & | GraphPt, |
| const TVec< TIntV > & | CmtyVVPt, | ||
| const TRnd & | RndPt | ||
| ) | [inline] |
| void TAGMFit::AddBaseCmty | ( | ) |
Add Epsilon community (base community which includes all nodes) into community affiliation graph. This means that we will later fit the value of epsilon.
Definition at line 251 of file agmfit.cpp.
References TVec< TVal, TSizeTy >::Add(), BaseCID, CIDNSetV, G, GetCmtyVV(), TUNGraph::GetNIdV(), IAssert, InitNodeData(), TVec< TVal, TSizeTy >::Len(), PNoCom, and SetCmtyVV().
{
TVec<TIntV> CmtyVV;
GetCmtyVV(CmtyVV);
TIntV TmpV = CmtyVV[0];
CmtyVV.Add(TmpV);
G->GetNIdV(CmtyVV[0]);
IAssert(CIDNSetV.Len() + 1 == CmtyVV.Len());
SetCmtyVV(CmtyVV);
InitNodeData();
BaseCID = 0;
PNoCom = 0.0;
}
| double TAGMFit::CalcPNoComByCmtyVV | ( | const int & | SamplePairs = -1 | ) |
Compute the empirical edge probability between a pair of nodes who share no community (epsilon), based on current community affiliations.
Definition at line 632 of file agmfit.cpp.
References G, THash< TKey, TDat, THashFunc >::GetDat(), TAGMUtil::GetIntersection(), TUNGraph::GetNIdV(), TUNGraph::GetNodes(), TUInt64::GetStr(), TUNGraph::IsEdge(), TVec< TVal, TSizeTy >::Len(), THashSet< TKey, THashFunc >::Len(), NIDComVH, PNoCom, and TFlt::Val.
{
TIntV NIdV;
G->GetNIdV(NIdV);
uint64 PairNoCom = 0, EdgesNoCom = 0;
for (int u = 0; u < NIdV.Len(); u++) {
for (int v = u + 1; v < NIdV.Len(); v++) {
int SrcNID = NIdV[u], DstNID = NIdV[v];
TIntSet JointCom;
TAGMUtil::GetIntersection(NIDComVH.GetDat(SrcNID),NIDComVH.GetDat(DstNID),JointCom);
if(JointCom.Len() == 0) {
PairNoCom++;
if (G->IsEdge(SrcNID, DstNID)) { EdgesNoCom++; }
if (SamplePairs > 0 && PairNoCom >= (uint64) SamplePairs) { break; }
}
}
if (SamplePairs > 0 && PairNoCom >= (uint64) SamplePairs) { break; }
}
double DefaultVal = 1.0 / (double)G->GetNodes() / (double)G->GetNodes();
if (EdgesNoCom > 0) {
PNoCom = (double) EdgesNoCom / (double) PairNoCom;
} else {
PNoCom = DefaultVal;
}
printf("%s / %s edges without joint com detected (PNoCom = %f)\n", TUInt64::GetStr(EdgesNoCom).CStr(), TUInt64::GetStr(PairNoCom).CStr(), PNoCom.Val);
return PNoCom;
}
| void TAGMFit::GetCmtyVV | ( | TVec< TIntV > & | CmtyVV, |
| const double | QMax = 2.0 |
||
| ) |
Get communities whose p_c is higher than 1 - QMax.
Definition at line 554 of file agmfit.cpp.
Referenced by AddBaseCmty(), and TAGMUtil::FindComsByAGM().

| void TAGMFit::GetCmtyVV | ( | TVec< TIntV > & | CmtyVV, |
| TFltV & | QV, | ||
| const double | QMax = 2.0 |
||
| ) |
COMMENT.
Definition at line 559 of file agmfit.cpp.
References TVec< TVal, TSizeTy >::Add(), THash< TKey, TDat, THashFunc >::AddDat(), BaseCID, CIDNSetV, G, TVec< TVal, TSizeTy >::Gen(), TUNGraph::GetNodes(), IAssert, LambdaV, TVec< TVal, TSizeTy >::Len(), and MinLambda.
{
CmtyVV.Gen(CIDNSetV.Len(), 0);
QV.Gen(CIDNSetV.Len(), 0);
TIntFltH CIDLambdaH(CIDNSetV.Len());
for (int c = 0; c < CIDNSetV.Len(); c++) {
CIDLambdaH.AddDat(c, LambdaV[c]);
}
CIDLambdaH.SortByDat(false);
for (int c = 0; c < CIDNSetV.Len(); c++) {
int CID = CIDLambdaH.GetKey(c);
IAssert(LambdaV[CID] >= MinLambda);
double Q = exp( - (double) LambdaV[CID]);
if (Q > QMax) { continue; }
TIntV CmtyV;
CIDNSetV[CID].GetKeyV(CmtyV);
if (CmtyV.Len() == 0) { continue; }
if (CID == BaseCID) { //if the community is the base community(epsilon community), discard
IAssert(CmtyV.Len() == G->GetNodes());
} else {
CmtyVV.Add(CmtyV);
QV.Add(Q);
}
}
}
| void TAGMFit::GetEdgeJointCom | ( | ) |
For each (u, v) in edges, precompute C_uv (the set of communities nodes u and v share).
Definition at line 50 of file agmfit.cpp.
References THash< TKey, TDat, THashFunc >::AddDat(), TUNGraph::BegNI(), CIDNSetV, ComEdgesV, EdgeComVH, TUNGraph::EndNI(), G, THash< TKey, TDat, THashFunc >::Gen(), TVec< TVal, TSizeTy >::Gen(), THash< TKey, TDat, THashFunc >::GetDat(), TUNGraph::GetEdges(), TAGMUtil::GetIntersection(), IAssert, THash< TKey, TDat, THashFunc >::IsKey(), THash< TKey, TDat, THashFunc >::Len(), TVec< TVal, TSizeTy >::Len(), THashSet< TKey, THashFunc >::Len(), and NIDComVH.
Referenced by InitNodeData().
{
ComEdgesV.Gen(CIDNSetV.Len());
EdgeComVH.Gen(G->GetEdges());
for (TUNGraph::TNodeI SrcNI = G->BegNI(); SrcNI < G->EndNI(); SrcNI++) {
int SrcNID = SrcNI.GetId();
for (int v = 0; v < SrcNI.GetDeg(); v++) {
int DstNID = SrcNI.GetNbrNId(v);
if (SrcNID >= DstNID) { continue; }
TIntSet JointCom;
IAssert(NIDComVH.IsKey(SrcNID));
IAssert(NIDComVH.IsKey(DstNID));
TAGMUtil::GetIntersection(NIDComVH.GetDat(SrcNID), NIDComVH.GetDat(DstNID), JointCom);
EdgeComVH.AddDat(TIntPr(SrcNID,DstNID),JointCom);
for (int k = 0; k < JointCom.Len(); k++) {
ComEdgesV[JointCom[k]]++;
}
}
}
IAssert(EdgeComVH.Len() == G->GetEdges());
}

| void TAGMFit::GetLambdaV | ( | TFltV & | OutV | ) | [inline] |
| void TAGMFit::GetNewtonStep | ( | TFltVV & | HVV, |
| TFltV & | GradV, | ||
| TFltV & | DeltaLV | ||
| ) |
| double TAGMFit::GetPNoCom | ( | ) | [inline] |
| void TAGMFit::GetQV | ( | TFltV & | OutV | ) |
Returns QV, a vector of (1 - p_c) for each community c.
Definition at line 451 of file agmfit.cpp.
References TVec< TVal, TSizeTy >::Gen(), LambdaV, and TVec< TVal, TSizeTy >::Len().
{
OutV.Gen(LambdaV.Len());
for (int i = 0; i < LambdaV.Len(); i++) {
OutV[i] = exp(- LambdaV[i]);
}
}
| double TAGMFit::GetStepSizeByLineSearchForLambda | ( | const TFltV & | DeltaV, |
| const TFltV & | GradV, | ||
| const double & | Alpha, | ||
| const double & | Beta | ||
| ) |
Step size search for updating P_c (which is parametarized by regularization parameter lambda).
Definition at line 109 of file agmfit.cpp.
References TLinAlg::DotProduct(), IAssert, LambdaV, TVec< TVal, TSizeTy >::Len(), Likelihood(), MaxLambda, and MinLambda.
Referenced by MLEGradAscentGivenCAG().
{
double StepSize = 1.0;
double InitLikelihood = Likelihood();
IAssert(LambdaV.Len() == DeltaV.Len());
TFltV NewLambdaV(LambdaV.Len());
for (int iter = 0; ; iter++) {
for (int i = 0; i < LambdaV.Len(); i++) {
NewLambdaV[i] = LambdaV[i] + StepSize * DeltaV[i];
if (NewLambdaV[i] < MinLambda) { NewLambdaV[i] = MinLambda; }
if (NewLambdaV[i] > MaxLambda) { NewLambdaV[i] = MaxLambda; }
}
if (Likelihood(NewLambdaV) < InitLikelihood + Alpha * StepSize * TLinAlg::DotProduct(GradV, DeltaV)) {
StepSize *= Beta;
} else {
break;
}
}
return StepSize;
}

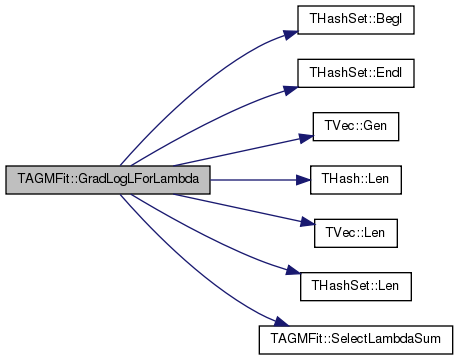
| void TAGMFit::GradLogLForLambda | ( | TFltV & | GradV | ) |
Definition at line 595 of file agmfit.cpp.
References THashSet< TKey, THashFunc >::BegI(), CIDNSetV, ComEdgesV, EdgeComVH, THashSet< TKey, THashFunc >::EndI(), TVec< TVal, TSizeTy >::Gen(), LambdaV, THash< TKey, TDat, THashFunc >::Len(), TVec< TVal, TSizeTy >::Len(), THashSet< TKey, THashFunc >::Len(), PNoCom, RegCoef, and SelectLambdaSum().
Referenced by MLEGradAscentGivenCAG().
{
GradV.Gen(LambdaV.Len());
TFltV SumEdgeProbsV(LambdaV.Len());
for (int e = 0; e < EdgeComVH.Len(); e++) {
TIntSet& JointCom = EdgeComVH[e];
double LambdaSum = SelectLambdaSum(JointCom);
double Puv = 1 - exp(- LambdaSum);
if (JointCom.Len() == 0) { Puv = PNoCom; }
for (TIntSet::TIter SI = JointCom.BegI(); SI < JointCom.EndI(); SI++) {
SumEdgeProbsV[SI.GetKey()] += (1 - Puv) / Puv;
}
}
for (int k = 0; k < LambdaV.Len(); k++) {
int MaxEk = CIDNSetV[k].Len() * (CIDNSetV[k].Len() - 1) / 2;
int NotEdgesInCom = MaxEk - ComEdgesV[k];
GradV[k] = SumEdgeProbsV[k] - (double) NotEdgesInCom;
if (LambdaV[k] > 0.0 && RegCoef > 0.0) { //if regularization exists
GradV[k] -= RegCoef;
}
}
}

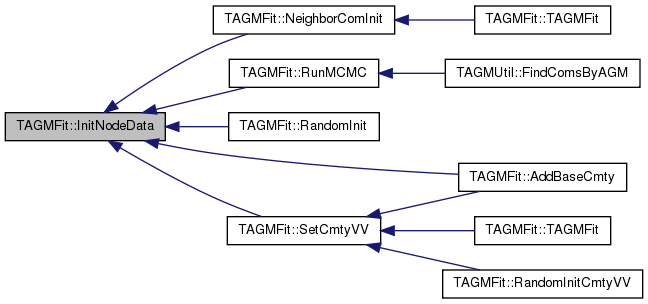
| void TAGMFit::InitNodeData | ( | ) |
COMMENT.
Definition at line 264 of file agmfit.cpp.
References THash< TKey, TDat, THashFunc >::AddDat(), THash< TKey, TDat, THashFunc >::AddKey(), TUNGraph::BegNI(), CIDNSetV, ComEdgesV, TSnap::DelSelfEdges(), TVec< TVal, TSizeTy >::EndI(), TUNGraph::EndNI(), G, THash< TKey, TDat, THashFunc >::Gen(), TVec< TVal, TSizeTy >::Gen(), GetEdgeJointCom(), TAGMUtil::GetNodeMembership(), TUNGraph::GetNodes(), LambdaV, TVec< TVal, TSizeTy >::Len(), MaxLambda, MinLambda, NIDCIDPrS, and NIDComVH.
Referenced by AddBaseCmty(), NeighborComInit(), RandomInit(), RunMCMC(), and SetCmtyVV().
{
TSnap::DelSelfEdges(G);
NIDComVH.Gen(G->GetNodes());
for (TUNGraph::TNodeI NI = G->BegNI(); NI < G->EndNI(); NI++) {
NIDComVH.AddDat(NI.GetId());
}
TAGMUtil::GetNodeMembership(NIDComVH, CIDNSetV);
GetEdgeJointCom();
LambdaV.Gen(CIDNSetV.Len());
for (int c = 0; c < CIDNSetV.Len(); c++) {
int MaxE = (CIDNSetV[c].Len()) * (CIDNSetV[c].Len() - 1) / 2;
if (MaxE < 2) {
LambdaV[c] = MaxLambda;
}
else{
LambdaV[c] = -log((double) (MaxE - ComEdgesV[c]) / MaxE);
}
if (LambdaV[c] > MaxLambda) { LambdaV[c] = MaxLambda; }
if (LambdaV[c] < MinLambda) { LambdaV[c] = MinLambda; }
}
NIDCIDPrS.Gen(G->GetNodes() * 10);
for (int c = 0; c < CIDNSetV.Len(); c++) {
for (TIntSet::TIter SI = CIDNSetV[c].BegI(); SI < CIDNSetV[c].EndI(); SI++) {
NIDCIDPrS.AddKey(TIntPr(SI.GetKey(), c));
}
}
}

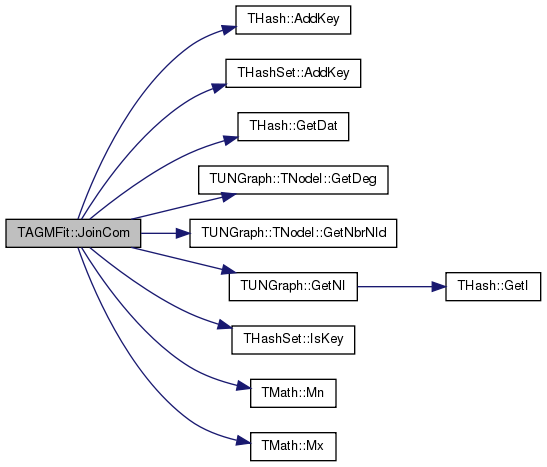
| void TAGMFit::JoinCom | ( | const int & | NID, |
| const int & | JoinCID | ||
| ) |
Definition at line 309 of file agmfit.cpp.
References THash< TKey, TDat, THashFunc >::AddKey(), THashSet< TKey, THashFunc >::AddKey(), CIDNSetV, ComEdgesV, EdgeComVH, G, THash< TKey, TDat, THashFunc >::GetDat(), TUNGraph::TNodeI::GetDeg(), TUNGraph::TNodeI::GetNbrNId(), TUNGraph::GetNI(), THashSet< TKey, THashFunc >::IsKey(), TMath::Mn(), TMath::Mx(), NIDCIDPrS, and NIDComVH.
Referenced by RunMCMC().
{
TUNGraph::TNodeI NI = G->GetNI(NID);
for (int e = 0; e < NI.GetDeg(); e++) {
int VID = NI.GetNbrNId(e);
if (NIDComVH.GetDat(VID).IsKey(JoinCID)) {
TIntPr SrcDstNIDPr = TIntPr(TMath::Mn(NID,VID), TMath::Mx(NID,VID));
EdgeComVH.GetDat(SrcDstNIDPr).AddKey(JoinCID);
ComEdgesV[JoinCID]++;
}
}
CIDNSetV[JoinCID].AddKey(NID);
NIDComVH.GetDat(NID).AddKey(JoinCID);
NIDCIDPrS.AddKey(TIntPr(NID, JoinCID));
}

| void TAGMFit::LeaveCom | ( | const int & | NID, |
| const int & | CID | ||
| ) |
After MCMC, NID leaves community CID.
Definition at line 293 of file agmfit.cpp.
References CIDNSetV, ComEdgesV, THash< TKey, TDat, THashFunc >::DelKey(), THashSet< TKey, THashFunc >::DelKey(), EdgeComVH, G, THash< TKey, TDat, THashFunc >::GetDat(), TUNGraph::TNodeI::GetDeg(), TUNGraph::TNodeI::GetNbrNId(), TUNGraph::GetNI(), THashSet< TKey, THashFunc >::IsKey(), TMath::Mn(), TMath::Mx(), NIDCIDPrS, and NIDComVH.
Referenced by RunMCMC().
{
TUNGraph::TNodeI NI = G->GetNI(NID);
for (int e = 0; e < NI.GetDeg(); e++) {
int VID = NI.GetNbrNId(e);
if (NIDComVH.GetDat(VID).IsKey(CID)) {
TIntPr SrcDstNIDPr = TIntPr(TMath::Mn(NID,VID), TMath::Mx(NID,VID));
EdgeComVH.GetDat(SrcDstNIDPr).DelKey(CID);
ComEdgesV[CID]--;
}
}
CIDNSetV[CID].DelKey(NID);
NIDComVH.GetDat(NID).DelKey(CID);
NIDCIDPrS.DelKey(TIntPr(NID, CID));
}


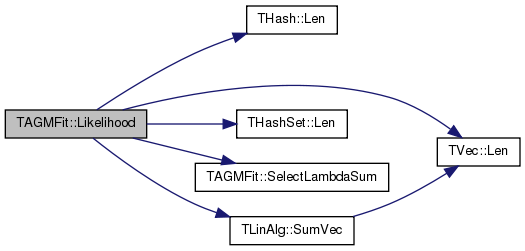
| double TAGMFit::Likelihood | ( | ) |
COMMENT.
Definition at line 104 of file agmfit.cpp.
References LambdaV.
Referenced by TAGMUtil::FindComsByAGM(), GetStepSizeByLineSearchForLambda(), MLEGradAscentGivenCAG(), PrintSummary(), and RunMCMC().
{
return Likelihood(LambdaV);
}

| double TAGMFit::Likelihood | ( | const TFltV & | NewLambdaV | ) | [inline] |
Definition at line 43 of file agmfit.h.
References Likelihood().
Referenced by Likelihood().
{ double Tmp1, Tmp2; return Likelihood(NewLambdaV, Tmp1, Tmp2); }


| double TAGMFit::Likelihood | ( | const TFltV & | NewLambdaV, |
| double & | LEdges, | ||
| double & | LNoEdges | ||
| ) |
Definition at line 76 of file agmfit.cpp.
References CIDNSetV, ComEdgesV, EdgeComVH, IAssert, THash< TKey, TDat, THashFunc >::Len(), TVec< TVal, TSizeTy >::Len(), THashSet< TKey, THashFunc >::Len(), TFlt::Mn, PNoCom, RegCoef, SelectLambdaSum(), and TLinAlg::SumVec().
{
IAssert(CIDNSetV.Len() == NewLambdaV.Len());
IAssert(ComEdgesV.Len() == CIDNSetV.Len());
LEdges = 0.0; LNoEdges = 0.0;
for (int e = 0; e < EdgeComVH.Len(); e++) {
TIntSet& JointCom = EdgeComVH[e];
double LambdaSum = SelectLambdaSum(NewLambdaV, JointCom);
double Puv = 1 - exp(- LambdaSum);
if (JointCom.Len() == 0) { Puv = PNoCom; }
IAssert(! _isnan(log(Puv)));
LEdges += log(Puv);
}
for (int k = 0; k < NewLambdaV.Len(); k++) {
int MaxEk = CIDNSetV[k].Len() * (CIDNSetV[k].Len() - 1) / 2;
int NotEdgesInCom = MaxEk - ComEdgesV[k];
if(NotEdgesInCom > 0) {
if (LNoEdges >= TFlt::Mn + double(NotEdgesInCom) * NewLambdaV[k]) {
LNoEdges -= double(NotEdgesInCom) * NewLambdaV[k];
}
}
}
double LReg = 0.0;
if (RegCoef > 0.0) {
LReg = - RegCoef * TLinAlg::SumVec(NewLambdaV);
}
return LEdges + LNoEdges + LReg;
}
| void TAGMFit::Load | ( | TSIn & | SIn, |
| const int & | RndSeed = 0 |
||
| ) |
Definition at line 24 of file agmfit.cpp.
References BaseCID, CIDNSetV, ComEdgesV, EdgeComVH, G, LambdaV, THash< TKey, TDat, THashFunc >::Load(), TVec< TVal, TSizeTy >::Load(), TInt::Load(), TFlt::Load(), MaxLambda, MinLambda, NIDCIDPrH, NIDCIDPrS, NIDComVH, PNoCom, TRnd::PutSeed(), RegCoef, and Rnd.
{
G = TUNGraph::Load(SIn);
CIDNSetV.Load(SIn);
EdgeComVH.Load(SIn);
NIDComVH.Load(SIn);
ComEdgesV.Load(SIn);
PNoCom.Load(SIn);
LambdaV.Load(SIn);
NIDCIDPrH.Load(SIn);
NIDCIDPrS.Load(SIn);
MinLambda.Load(SIn);
MaxLambda.Load(SIn);
RegCoef.Load(SIn);
BaseCID.Load(SIn);
Rnd.PutSeed(RndSeed);
}
| int TAGMFit::MLEGradAscentGivenCAG | ( | const double & | Thres = 0.001, |
| const int & | MaxIter = 10000, |
||
| const TStr | PlotNm = TStr() |
||
| ) |
Gradient descent for p_c while keeping the community affiliation graph (CAG) fixed.
Definition at line 130 of file agmfit.cpp.
References TVec< TVal, TSizeTy >::Add(), TStr::Empty(), G, TUNGraph::GetEdges(), GetStepSizeByLineSearchForLambda(), TExeTm::GetTmStr(), GradLogLForLambda(), Kilo, LambdaV, TVec< TVal, TSizeTy >::Len(), Likelihood(), MaxLambda, MinLambda, TLinAlg::Norm(), and TGnuPlot::PlotValV().
Referenced by TAGMUtil::FindComsByAGM(), and RunMCMC().
{
int Edges = G->GetEdges();
TExeTm ExeTm;
TFltV GradV(LambdaV.Len());
int iter = 0;
TIntFltPrV IterLV, IterGradNormV;
double GradCutOff = 1000;
for (iter = 0; iter < MaxIter; iter++) {
GradLogLForLambda(GradV); //if gradient is going out of the boundary, cut off
for (int i = 0; i < LambdaV.Len(); i++) {
if (GradV[i] < -GradCutOff) { GradV[i] = -GradCutOff; }
if (GradV[i] > GradCutOff) { GradV[i] = GradCutOff; }
if (LambdaV[i] <= MinLambda && GradV[i] < 0) { GradV[i] = 0.0; }
if (LambdaV[i] >= MaxLambda && GradV[i] > 0) { GradV[i] = 0.0; }
}
double Alpha = 0.15, Beta = 0.2;
if (Edges > Kilo(100)) { Alpha = 0.00015; Beta = 0.3;}
double LearnRate = GetStepSizeByLineSearchForLambda(GradV, GradV, Alpha, Beta);
if (TLinAlg::Norm(GradV) < Thres) { break; }
for (int i = 0; i < LambdaV.Len(); i++) {
double Change = LearnRate * GradV[i];
LambdaV[i] += Change;
if(LambdaV[i] < MinLambda) { LambdaV[i] = MinLambda;}
if(LambdaV[i] > MaxLambda) { LambdaV[i] = MaxLambda;}
}
if (! PlotNm.Empty()) {
double L = Likelihood();
IterLV.Add(TIntFltPr(iter, L));
IterGradNormV.Add(TIntFltPr(iter, TLinAlg::Norm(GradV)));
}
}
if (! PlotNm.Empty()) {
TGnuPlot::PlotValV(IterLV, PlotNm + ".likelihood_Q");
TGnuPlot::PlotValV(IterGradNormV, PlotNm + ".gradnorm_Q");
printf("MLE for Lambda completed with %d iterations(%s)\n",iter,ExeTm.GetTmStr());
}
return iter;
}


| void TAGMFit::NeighborComInit | ( | const int | InitComs | ) |
Initialize node community memberships using best neighborhood communities (see D. Gleich et al. KDD'12).
Definition at line 186 of file agmfit.cpp.
References CIDNSetV, G, TVec< TVal, TSizeTy >::Gen(), TAGMUtil::GetConductance(), TUNGraph::TNodeI::GetDeg(), TUNGraph::GetEdges(), TUNGraph::TNodeI::GetId(), TAGMUtil::GetNbhCom(), TUNGraph::TNodeI::GetNbrNId(), TUNGraph::GetNI(), TUNGraph::GetNIdV(), TUNGraph::GetNodes(), TUNGraph::GetRndNI(), TExeTm::GetTmStr(), IAssert, InitNodeData(), TVec< TVal, TSizeTy >::Len(), and SetDefaultPNoCom().
Referenced by TAGMFit().
{
CIDNSetV.Gen(InitComs);
const int Edges = G->GetEdges();
TFltIntPrV NIdPhiV(G->GetNodes(), 0);
TIntSet InvalidNIDS(G->GetNodes());
TIntV ChosenNIDV(InitComs, 0); //FOR DEBUG
TExeTm RunTm;
//compute conductance of neighborhood community
TIntV NIdV;
G->GetNIdV(NIdV);
for (int u = 0; u < NIdV.Len(); u++) {
TIntSet NBCmty(G->GetNI(NIdV[u]).GetDeg() + 1);
double Phi;
if (G->GetNI(NIdV[u]).GetDeg() < 5) { //do not include nodes with too few degree
Phi = 1.0;
} else {
TAGMUtil::GetNbhCom(G, NIdV[u], NBCmty);
IAssert(NBCmty.Len() == G->GetNI(NIdV[u]).GetDeg() + 1);
Phi = TAGMUtil::GetConductance(G, NBCmty, Edges);
}
NIdPhiV.Add(TFltIntPr(Phi, NIdV[u]));
}
NIdPhiV.Sort(true);
printf("conductance computation completed [%s]\n", RunTm.GetTmStr());
fflush(stdout);
//choose nodes with local minimum in conductance
int CurCID = 0;
for (int ui = 0; ui < NIdPhiV.Len(); ui++) {
int UID = NIdPhiV[ui].Val2;
fflush(stdout);
if (InvalidNIDS.IsKey(UID)) { continue; }
ChosenNIDV.Add(UID); //FOR DEBUG
//add the node and its neighbors to the current community
CIDNSetV[CurCID].AddKey(UID);
TUNGraph::TNodeI NI = G->GetNI(UID);
fflush(stdout);
for (int e = 0; e < NI.GetDeg(); e++) {
CIDNSetV[CurCID].AddKey(NI.GetNbrNId(e));
}
//exclude its neighbors from the next considerations
for (int e = 0; e < NI.GetDeg(); e++) {
InvalidNIDS.AddKey(NI.GetNbrNId(e));
}
CurCID++;
fflush(stdout);
if (CurCID >= InitComs) { break; }
}
if (InitComs > CurCID) {
printf("%d communities needed to fill randomly\n", InitComs - CurCID);
}
//assign a member to zero-member community (if any)
for (int c = 0; c < CIDNSetV.Len(); c++) {
if (CIDNSetV[c].Len() == 0) {
int ComSz = 10;
for (int u = 0; u < ComSz; u++) {
int UID = G->GetRndNI().GetId();
CIDNSetV[c].AddKey(UID);
}
}
}
InitNodeData();
SetDefaultPNoCom();
}
| void TAGMFit::PrintSummary | ( | ) |
COMMENT.
Definition at line 659 of file agmfit.cpp.
References THash< TKey, TDat, THashFunc >::AddDat(), CIDNSetV, ComEdgesV, LambdaV, THash< TKey, TDat, THashFunc >::Len(), TVec< TVal, TSizeTy >::Len(), Likelihood(), NIDCIDPrS, PNoCom, and TFlt::Val.
{
TIntFltH CIDLambdaH(CIDNSetV.Len());
for (int c = 0; c < CIDNSetV.Len(); c++) {
CIDLambdaH.AddDat(c, LambdaV[c]);
}
CIDLambdaH.SortByDat(false);
int Coms = 0;
for (int i = 0; i < LambdaV.Len(); i++) {
int CID = CIDLambdaH.GetKey(i);
if (LambdaV[CID] <= 0.0001) { continue; }
printf("P_c : %.3f Com Sz: %d, Total Edges inside: %d \n", 1.0 - exp(- LambdaV[CID]), CIDNSetV[CID].Len(), (int) ComEdgesV[CID]);
Coms++;
}
printf("%d Communities, Total Memberships = %d, Likelihood = %.2f, Epsilon = %f\n", Coms, NIDCIDPrS.Len(), Likelihood(), PNoCom.Val);
}
| void TAGMFit::RandomInit | ( | const int & | MaxK | ) |
COMMENT.
Definition at line 169 of file agmfit.cpp.
References TVec< TVal, TSizeTy >::Add(), THashSet< TKey, THashFunc >::AddKey(), CIDNSetV, TVec< TVal, TSizeTy >::Clr(), G, TUNGraph::TNodeI::GetDeg(), TUNGraph::TNodeI::GetId(), TUNGraph::TNodeI::GetNbrNId(), TUNGraph::GetNI(), TRnd::GetUniDevInt(), InitNodeData(), TVec< TVal, TSizeTy >::Last(), Rnd, and SetDefaultPNoCom().
{
CIDNSetV.Clr();
for (int c = 0; c < MaxK; c++) {
CIDNSetV.Add();
int NC = Rnd.GetUniDevInt(G -> GetNodes());
TUNGraph::TNodeI NI = G -> GetRndNI();
CIDNSetV.Last().AddKey(NI.GetId());
for (int v = 0; v < NC; v++) {
NI = G->GetNI(NI.GetNbrNId(Rnd.GetUniDevInt(NI.GetDeg())));
CIDNSetV.Last().AddKey(NI.GetId());
}
}
InitNodeData();
SetDefaultPNoCom();
}

| void TAGMFit::RandomInitCmtyVV | ( | const int | InitComs, |
| const double | ComSzAlpha = 1.3, |
||
| const double | MemAlpha = 1.8, |
||
| const int | MinComSz = 8, |
||
| const int | MaxComSz = 200, |
||
| const int | MinMem = 1, |
||
| const int | MaxMem = 10 |
||
| ) |
Randomly initialize bipartite community affiliation graph.
Definition at line 42 of file agmfit.cpp.
References G, TAGMUtil::GenCmtyVVFromPL(), TUNGraph::GetNodes(), Rnd, and SetCmtyVV().
{
TVec<TIntV> InitCmtyVV(InitComs, 0);
TAGMUtil::GenCmtyVVFromPL(InitCmtyVV, G, G->GetNodes(), InitComs, ComSzAlpha, MemAlpha, MinComSz, MaxComSz,
MinMem, MaxMem, Rnd);
SetCmtyVV(InitCmtyVV);
}
| int TAGMFit::RemoveEmptyCom | ( | ) |
Remove all communities with no members.
Definition at line 459 of file agmfit.cpp.
References CIDNSetV, TVec< TVal, TSizeTy >::DelLast(), LambdaV, TVec< TVal, TSizeTy >::Last(), and TVec< TVal, TSizeTy >::Len().
{
int DelCnt = 0;
for (int c = 0; c < CIDNSetV.Len(); c++) {
if (CIDNSetV[c].Len() == 0) {
CIDNSetV[c] = CIDNSetV.Last();
CIDNSetV.DelLast();
LambdaV[c] = LambdaV.Last();
LambdaV.DelLast();
DelCnt++;
c--;
}
}
return DelCnt;
}
| void TAGMFit::RunMCMC | ( | const int & | MaxIter, |
| const int & | EvalLambdaIter, | ||
| const TStr & | PlotFPrx = TStr() |
||
| ) |
Main procedure for fitting the AGM to a given graph using MCMC.
MCMC fitting.
Definition at line 366 of file agmfit.cpp.
References TVec< TVal, TSizeTy >::Add(), TGnuPlot::AddPlot(), BaseCID, CIDNSetV, TStr::Fmt(), G, TUNGraph::GetEdges(), TUNGraph::GetNodes(), TExeTm::GetTmStr(), TRnd::GetUniDev(), gpwLinesPoints, InitNodeData(), JoinCom(), LambdaV, LeaveCom(), TStr::Len(), TVec< TVal, TSizeTy >::Len(), Likelihood(), MLEGradAscentGivenCAG(), TMath::Mx(), Rnd, SampleTransition(), TGnuPlot::SavePng(), TGnuPlot::SetDataPlotFNm(), TGnuPlot::SetTitle(), and TExeTm::Tick().
Referenced by TAGMUtil::FindComsByAGM().
{
TExeTm IterTm, TotalTm;
double PrevL = Likelihood(), DeltaL = 0;
double BestL = PrevL;
printf("initial likelihood = %f\n",PrevL);
TIntFltPrV IterTrueLV, IterJoinV, IterLeaveV, IterAcceptV, IterSwitchV, IterLBV;
TIntPrV IterTotMemV;
TIntV IterV;
TFltV BestLV;
TVec<TIntSet> BestCmtySetV;
int SwitchCnt = 0, LeaveCnt = 0, JoinCnt = 0, AcceptCnt = 0, ProbBinSz;
int Nodes = G->GetNodes(), Edges = G->GetEdges();
TExeTm PlotTm;
ProbBinSz = TMath::Mx(1000, G->GetNodes() / 10); //bin to compute probabilities
IterLBV.Add(TIntFltPr(1, BestL));
for (int iter = 0; iter < MaxIter; iter++) {
IterTm.Tick();
int NID = -1;
int JoinCID = -1, LeaveCID = -1;
SampleTransition(NID, JoinCID, LeaveCID, DeltaL); //sample a move
double OptL = PrevL;
if (DeltaL > 0 || Rnd.GetUniDev() < exp(DeltaL)) { //if it is accepted
IterTm.Tick();
if (LeaveCID > -1 && LeaveCID != BaseCID) { LeaveCom(NID, LeaveCID); }
if (JoinCID > -1 && JoinCID != BaseCID) { JoinCom(NID, JoinCID); }
if (LeaveCID > -1 && JoinCID > -1 && JoinCID != BaseCID && LeaveCID != BaseCID) { SwitchCnt++; }
else if (LeaveCID > -1 && LeaveCID != BaseCID) { LeaveCnt++;}
else if (JoinCID > -1 && JoinCID != BaseCID) { JoinCnt++;}
AcceptCnt++;
if ((iter + 1) % EvalLambdaIter == 0) {
IterTm.Tick();
MLEGradAscentGivenCAG(0.01, 3);
OptL = Likelihood();
}
else{
OptL = PrevL + DeltaL;
}
if (BestL <= OptL && CIDNSetV.Len() > 0) {
BestCmtySetV = CIDNSetV;
BestLV = LambdaV;
BestL = OptL;
}
}
if (iter > 0 && (iter % ProbBinSz == 0) && PlotFPrx.Len() > 0) {
IterLBV.Add(TIntFltPr(iter, OptL));
IterSwitchV.Add(TIntFltPr(iter, (double) SwitchCnt / (double) AcceptCnt));
IterLeaveV.Add(TIntFltPr(iter, (double) LeaveCnt / (double) AcceptCnt));
IterJoinV.Add(TIntFltPr(iter, (double) JoinCnt / (double) AcceptCnt));
IterAcceptV.Add(TIntFltPr(iter, (double) AcceptCnt / (double) ProbBinSz));
SwitchCnt = JoinCnt = LeaveCnt = AcceptCnt = 0;
}
PrevL = OptL;
if ((iter + 1) % 10000 == 0) {
printf("\r%d iterations completed [%.2f]", iter, (double) iter / (double) MaxIter);
}
}
// plot the likelihood and acceptance probabilities if the plot file name is given
if (PlotFPrx.Len() > 0) {
TGnuPlot GP1;
GP1.AddPlot(IterLBV, gpwLinesPoints, "likelihood");
GP1.SetDataPlotFNm(PlotFPrx + ".likelihood.tab", PlotFPrx + ".likelihood.plt");
TStr TitleStr = TStr::Fmt(" N:%d E:%d", Nodes, Edges);
GP1.SetTitle(PlotFPrx + ".likelihood" + TitleStr);
GP1.SavePng(PlotFPrx + ".likelihood.png");
TGnuPlot GP2;
GP2.AddPlot(IterSwitchV, gpwLinesPoints, "Switch");
GP2.AddPlot(IterLeaveV, gpwLinesPoints, "Leave");
GP2.AddPlot(IterJoinV, gpwLinesPoints, "Join");
GP2.AddPlot(IterAcceptV, gpwLinesPoints, "Accept");
GP2.SetTitle(PlotFPrx + ".transition");
GP2.SetDataPlotFNm(PlotFPrx + "transition_prob.tab", PlotFPrx + "transition_prob.plt");
GP2.SavePng(PlotFPrx + "transition_prob.png");
}
CIDNSetV = BestCmtySetV;
LambdaV = BestLV;
InitNodeData();
MLEGradAscentGivenCAG(0.001, 100);
printf("\nMCMC completed (best likelihood: %.2f) [%s]\n", BestL, TotalTm.GetTmStr());
}


| void TAGMFit::SampleTransition | ( | int & | NID, |
| int & | JoinCID, | ||
| int & | LeaveCID, | ||
| double & | DeltaL | ||
| ) |
Sample MMCM transitions: Choose among (join, leave, switch), and then sample (NID, CID).
Definition at line 325 of file agmfit.cpp.
References BaseCID, CIDNSetV, G, THash< TKey, TDat, THashFunc >::GetDat(), THash< TKey, TDat, THashFunc >::GetKey(), TUNGraph::GetNodes(), THash< TKey, TDat, THashFunc >::GetRndKeyId(), TUNGraph::GetRndNId(), TRnd::GetUniDevInt(), THash< TKey, TDat, THashFunc >::IsKey(), THash< TKey, TDat, THashFunc >::Len(), TVec< TVal, TSizeTy >::Len(), THashSet< TKey, THashFunc >::Len(), TFlt::Mn, NIDCIDPrS, NIDComVH, Rnd, SeekJoin(), SeekLeave(), SeekSwitch(), TPair< TVal1, TVal2 >::Val1, and TPair< TVal1, TVal2 >::Val2.
Referenced by RunMCMC().
{
int Option = Rnd.GetUniDevInt(3); //0:Join 1:Leave 2:Switch
if (NIDCIDPrS.Len() <= 1) { Option = 0; } //if there is only one node membership, only join is possible.
int TryCnt = 0;
const int MaxTryCnt = G->GetNodes();
DeltaL = TFlt::Mn;
if (Option == 0) {
do {
JoinCID = Rnd.GetUniDevInt(CIDNSetV.Len());
NID = G->GetRndNId();
} while (TryCnt++ < MaxTryCnt && NIDCIDPrS.IsKey(TIntPr(NID, JoinCID)));
if (TryCnt < MaxTryCnt) { //if successfully find a move
DeltaL = SeekJoin(NID, JoinCID);
}
}
else if (Option == 1) {
do {
TIntPr NIDCIDPr = NIDCIDPrS.GetKey(NIDCIDPrS.GetRndKeyId(Rnd));
NID = NIDCIDPr.Val1;
LeaveCID = NIDCIDPr.Val2;
} while (TryCnt++ < MaxTryCnt && LeaveCID == BaseCID);
if (TryCnt < MaxTryCnt) {//if successfully find a move
DeltaL = SeekLeave(NID, LeaveCID);
}
}
else{
do {
TIntPr NIDCIDPr = NIDCIDPrS.GetKey(NIDCIDPrS.GetRndKeyId(Rnd));
NID = NIDCIDPr.Val1;
LeaveCID = NIDCIDPr.Val2;
} while (TryCnt++ < MaxTryCnt && (NIDComVH.GetDat(NID).Len() == CIDNSetV.Len() || LeaveCID == BaseCID));
do {
JoinCID = Rnd.GetUniDevInt(CIDNSetV.Len());
} while (TryCnt++ < G->GetNodes() && NIDCIDPrS.IsKey(TIntPr(NID, JoinCID)));
if (TryCnt < MaxTryCnt) {//if successfully find a move
DeltaL = SeekSwitch(NID, LeaveCID, JoinCID);
}
}
}

| void TAGMFit::Save | ( | TSOut & | SOut | ) |
Definition at line 8 of file agmfit.cpp.
References BaseCID, CIDNSetV, ComEdgesV, EdgeComVH, G, LambdaV, MaxLambda, MinLambda, NIDCIDPrH, NIDCIDPrS, NIDComVH, PNoCom, RegCoef, THash< TKey, TDat, THashFunc >::Save(), TUNGraph::Save(), TVec< TVal, TSizeTy >::Save(), TInt::Save(), and TFlt::Save().
{
G->Save(SOut);
CIDNSetV.Save(SOut);
EdgeComVH.Save(SOut);
NIDComVH.Save(SOut);
ComEdgesV.Save(SOut);
PNoCom.Save(SOut);
LambdaV.Save(SOut);
NIDCIDPrH.Save(SOut);
NIDCIDPrS.Save(SOut);
MinLambda.Save(SOut);
MaxLambda.Save(SOut);
RegCoef.Save(SOut);
BaseCID.Save(SOut);
}
| double TAGMFit::SeekJoin | ( | const int & | UID, |
| const int & | CID | ||
| ) |
Compute the change in likelihood (Delta) if node UID joins community CID.
Definition at line 502 of file agmfit.cpp.
References CIDNSetV, EdgeComVH, G, THash< TKey, TDat, THashFunc >::GetDat(), TUNGraph::TNodeI::GetDeg(), TUNGraph::TNodeI::GetNbrNId(), TUNGraph::GetNI(), IAssert, THashSet< TKey, THashFunc >::IsKey(), LambdaV, TVec< TVal, TSizeTy >::Len(), THashSet< TKey, THashFunc >::Len(), TMath::Mn(), TMath::Mx(), NIDComVH, PNoCom, and SelectLambdaSum().
Referenced by SampleTransition(), and SeekSwitch().
{
IAssert(! CIDNSetV[CID].IsKey(UID));
double Delta = 0.0;
TUNGraph::TNodeI NI = G->GetNI(UID);
int NbhsInC = 0;
for (int e = 0; e < NI.GetDeg(); e++) {
const int VID = NI.GetNbrNId(e);
if (! NIDComVH.GetDat(VID).IsKey(CID)) { continue; }
TIntPr SrcDstNIDPr(TMath::Mn(UID,VID), TMath::Mx(UID,VID));
TIntSet& JointCom = EdgeComVH.GetDat(SrcDstNIDPr);
double CurPuv, NewPuv, LambdaSum = SelectLambdaSum(JointCom);
CurPuv = 1 - exp(- LambdaSum);
if (JointCom.Len() == 0) { CurPuv = PNoCom; }
NewPuv = 1 - exp(- LambdaSum - LambdaV[CID]);
Delta += (log(NewPuv) - log(CurPuv));
IAssert(!_isnan(Delta));
NbhsInC++;
}
Delta -= LambdaV[CID] * (CIDNSetV[CID].Len() - NbhsInC);
return Delta;
}

| double TAGMFit::SeekLeave | ( | const int & | UID, |
| const int & | CID | ||
| ) |
Compute the change in likelihood (Delta) if node UID leaves community CID.
Definition at line 475 of file agmfit.cpp.
References CIDNSetV, EdgeComVH, G, THash< TKey, TDat, THashFunc >::GetDat(), TUNGraph::TNodeI::GetDeg(), TUNGraph::TNodeI::GetNbrNId(), TUNGraph::GetNI(), IAssert, THashSet< TKey, THashFunc >::IsKey(), TUNGraph::IsNode(), LambdaV, TVec< TVal, TSizeTy >::Len(), THashSet< TKey, THashFunc >::Len(), TMath::Mn(), TMath::Mx(), NIDComVH, PNoCom, and SelectLambdaSum().
Referenced by SampleTransition(), and SeekSwitch().
{
IAssert(CIDNSetV[CID].IsKey(UID));
IAssert(G->IsNode(UID));
double Delta = 0.0;
TUNGraph::TNodeI NI = G->GetNI(UID);
int NbhsInC = 0;
for (int e = 0; e < NI.GetDeg(); e++) {
const int VID = NI.GetNbrNId(e);
if (! NIDComVH.GetDat(VID).IsKey(CID)) { continue; }
TIntPr SrcDstNIDPr(TMath::Mn(UID,VID), TMath::Mx(UID,VID));
TIntSet& JointCom = EdgeComVH.GetDat(SrcDstNIDPr);
double CurPuv, NewPuv, LambdaSum = SelectLambdaSum(JointCom);
CurPuv = 1 - exp(- LambdaSum);
NewPuv = 1 - exp(- LambdaSum + LambdaV[CID]);
IAssert(JointCom.Len() > 0);
if (JointCom.Len() == 1) {
NewPuv = PNoCom;
}
Delta += (log(NewPuv) - log(CurPuv));
IAssert(!_isnan(Delta));
NbhsInC++;
}
Delta += LambdaV[CID] * (CIDNSetV[CID].Len() - 1 - NbhsInC);
return Delta;
}

| double TAGMFit::SeekSwitch | ( | const int & | UID, |
| const int & | CurCID, | ||
| const int & | NewCID | ||
| ) |
Definition at line 525 of file agmfit.cpp.
References CIDNSetV, EdgeComVH, G, THash< TKey, TDat, THashFunc >::GetDat(), TUNGraph::TNodeI::GetDeg(), TUNGraph::TNodeI::GetNbrNId(), TUNGraph::GetNI(), IAssert, THashSet< TKey, THashFunc >::IsKey(), LambdaV, THashSet< TKey, THashFunc >::Len(), TMath::Mn(), TMath::Mx(), NIDComVH, PNoCom, SeekJoin(), SeekLeave(), SelectLambdaSum(), and TFlt::Val.
Referenced by SampleTransition().
{
IAssert(! CIDNSetV[NewCID].IsKey(UID));
IAssert(CIDNSetV[CurCID].IsKey(UID));
double Delta = SeekJoin(UID, NewCID) + SeekLeave(UID, CurCID);
//correct only for intersection between new com and current com
TUNGraph::TNodeI NI = G->GetNI(UID);
for (int e = 0; e < NI.GetDeg(); e++) {
const int VID = NI.GetNbrNId(e);
if (! NIDComVH.GetDat(VID).IsKey(CurCID) || ! NIDComVH.GetDat(VID).IsKey(NewCID)) {continue;}
TIntPr SrcDstNIDPr(TMath::Mn(UID,VID), TMath::Mx(UID,VID));
TIntSet& JointCom = EdgeComVH.GetDat(SrcDstNIDPr);
double CurPuv, NewPuvAfterJoin, NewPuvAfterLeave, NewPuvAfterSwitch, LambdaSum = SelectLambdaSum(JointCom);
CurPuv = 1 - exp(- LambdaSum);
NewPuvAfterLeave = 1 - exp(- LambdaSum + LambdaV[CurCID]);
NewPuvAfterJoin = 1 - exp(- LambdaSum - LambdaV[NewCID]);
NewPuvAfterSwitch = 1 - exp(- LambdaSum - LambdaV[NewCID] + LambdaV[CurCID]);
if (JointCom.Len() == 1 || NewPuvAfterLeave == 0.0) {
NewPuvAfterLeave = PNoCom;
}
Delta += (log(NewPuvAfterSwitch) + log(CurPuv) - log(NewPuvAfterLeave) - log(NewPuvAfterJoin));
if (_isnan(Delta)) {
printf("NS:%f C:%f NL:%f NJ:%f PNoCom:%f", NewPuvAfterSwitch, CurPuv, NewPuvAfterLeave, NewPuvAfterJoin, PNoCom.Val);
}
IAssert(!_isnan(Delta));
}
return Delta;
}

| double TAGMFit::SelectLambdaSum | ( | const TIntSet & | ComK | ) |
Compute sum of lambda_c (which is log (1 - p_c)) over C_uv (ComK). The function is used to compute edge probability P_uv.
Definition at line 618 of file agmfit.cpp.
References LambdaV.
Referenced by GradLogLForLambda(), Likelihood(), SeekJoin(), SeekLeave(), and SeekSwitch().
{
return SelectLambdaSum(LambdaV, ComK);
}
| double TAGMFit::SelectLambdaSum | ( | const TFltV & | NewLambdaV, |
| const TIntSet & | ComK | ||
| ) |
COMMENT.
Definition at line 622 of file agmfit.cpp.
References THashSet< TKey, THashFunc >::BegI(), THashSet< TKey, THashFunc >::EndI(), and IAssert.
{
double Result = 0.0;
for (TIntSet::TIter SI = ComK.BegI(); SI < ComK.EndI(); SI++) {
IAssert(NewLambdaV[SI.GetKey()] >= 0);
Result += NewLambdaV[SI.GetKey()];
}
return Result;
}

| void TAGMFit::SetCmtyVV | ( | const TVec< TIntV > & | CmtyVV | ) |
COMMENT.
Definition at line 584 of file agmfit.cpp.
References CIDNSetV, G, TVec< TVal, TSizeTy >::Gen(), IAssert, InitNodeData(), TUNGraph::IsNode(), TVec< TVal, TSizeTy >::Len(), and SetDefaultPNoCom().
Referenced by AddBaseCmty(), RandomInitCmtyVV(), and TAGMFit().
{
CIDNSetV.Gen(CmtyVV.Len());
for (int c = 0; c < CIDNSetV.Len(); c++) {
CIDNSetV[c].AddKeyV(CmtyVV[c]);
for (int j = 0; j < CmtyVV[c].Len(); j++) { IAssert(G->IsNode(CmtyVV[c][j])); } // check whether the member nodes exist in the graph
}
InitNodeData();
SetDefaultPNoCom();
}

| void TAGMFit::SetDefaultPNoCom | ( | ) |
Set Epsilon (the probability that two nodes sharing no communities connect) to the default value.
Definition at line 72 of file agmfit.cpp.
References G, TUNGraph::GetNodes(), and PNoCom.
Referenced by NeighborComInit(), RandomInit(), and SetCmtyVV().
| void TAGMFit::SetLambdaV | ( | const TFltV & | LambdaPt | ) | [inline] |
| void TAGMFit::SetPNoCom | ( | const double & | Epsilon | ) | [inline] |
Set Epsilon (the probability that two nodes sharing no communities connect) to the default value.
Definition at line 57 of file agmfit.h.
References BaseCID, and PNoCom.
Referenced by TAGMUtil::FindComsByAGM().

| void TAGMFit::SetRegCoef | ( | const double | Val | ) | [inline] |
Definition at line 45 of file agmfit.h.
References RegCoef.
Referenced by TAGMUtil::FindComsByAGM().
{ RegCoef = Val; }

TInt TAGMFit::BaseCID [private] |
ID of the Epsilon-community (in case we fit P_c of the epsilon community). We do not fit for the Epsilon-community in general.
Definition at line 23 of file agmfit.h.
Referenced by AddBaseCmty(), GetCmtyVV(), Load(), RunMCMC(), SampleTransition(), Save(), and SetPNoCom().
TVec<TIntSet> TAGMFit::CIDNSetV [private] |
Community ID -> Member Node ID Sets.
Definition at line 11 of file agmfit.h.
Referenced by AddBaseCmty(), GetCmtyVV(), GetEdgeJointCom(), GradLogLForLambda(), InitNodeData(), JoinCom(), LeaveCom(), Likelihood(), Load(), NeighborComInit(), PrintSummary(), RandomInit(), RemoveEmptyCom(), RunMCMC(), SampleTransition(), Save(), SeekJoin(), SeekLeave(), SeekSwitch(), and SetCmtyVV().
TIntV TAGMFit::ComEdgesV [private] |
The number of edges in each community.
Definition at line 14 of file agmfit.h.
Referenced by GetEdgeJointCom(), GradLogLForLambda(), InitNodeData(), JoinCom(), LeaveCom(), Likelihood(), Load(), PrintSummary(), and Save().
THash<TIntPr,TIntSet> TAGMFit::EdgeComVH [private] |
Edge -> Shared Community ID Set.
Definition at line 12 of file agmfit.h.
Referenced by GetEdgeJointCom(), GradLogLForLambda(), JoinCom(), LeaveCom(), Likelihood(), Load(), Save(), SeekJoin(), SeekLeave(), and SeekSwitch().
PUNGraph TAGMFit::G [private] |
Graph to fit.
Definition at line 10 of file agmfit.h.
Referenced by AddBaseCmty(), CalcPNoComByCmtyVV(), GetCmtyVV(), GetEdgeJointCom(), InitNodeData(), JoinCom(), LeaveCom(), Load(), MLEGradAscentGivenCAG(), NeighborComInit(), RandomInit(), RandomInitCmtyVV(), RunMCMC(), SampleTransition(), Save(), SeekJoin(), SeekLeave(), SeekSwitch(), SetCmtyVV(), and SetDefaultPNoCom().
TFltV TAGMFit::LambdaV [private] |
Parametrization of P_c (edge probability in community c), P_c = 1 - exp(-lambda).
Definition at line 16 of file agmfit.h.
Referenced by GetCmtyVV(), GetLambdaV(), GetQV(), GetStepSizeByLineSearchForLambda(), GradLogLForLambda(), InitNodeData(), Likelihood(), Load(), MLEGradAscentGivenCAG(), PrintSummary(), RemoveEmptyCom(), RunMCMC(), Save(), SeekJoin(), SeekLeave(), SeekSwitch(), SelectLambdaSum(), and SetLambdaV().
TFlt TAGMFit::MaxLambda [private] |
Maximum value of regularization parameter lambda (default = 10).
Definition at line 21 of file agmfit.h.
Referenced by GetStepSizeByLineSearchForLambda(), InitNodeData(), Load(), MLEGradAscentGivenCAG(), and Save().
TFlt TAGMFit::MinLambda [private] |
Minimum value of regularization parameter lambda (default = 1e-5).
Definition at line 20 of file agmfit.h.
Referenced by GetCmtyVV(), GetStepSizeByLineSearchForLambda(), InitNodeData(), Load(), MLEGradAscentGivenCAG(), and Save().
THash<TIntPr,TFlt> TAGMFit::NIDCIDPrH [private] |
THash<TIntPr,TInt> TAGMFit::NIDCIDPrS [private] |
<Node ID, Community ID> pairs (for sampling MCMC moves).
Definition at line 19 of file agmfit.h.
Referenced by InitNodeData(), JoinCom(), LeaveCom(), Load(), PrintSummary(), SampleTransition(), and Save().
THash<TInt, TIntSet> TAGMFit::NIDComVH [private] |
Node ID -> Communitie IDs the node belongs to.
Definition at line 13 of file agmfit.h.
Referenced by CalcPNoComByCmtyVV(), GetEdgeJointCom(), InitNodeData(), JoinCom(), LeaveCom(), Load(), SampleTransition(), Save(), SeekJoin(), SeekLeave(), and SeekSwitch().
TFlt TAGMFit::PNoCom [private] |
Probability of edge when two nodes share no community (epsilon in the paper).
Definition at line 15 of file agmfit.h.
Referenced by AddBaseCmty(), CalcPNoComByCmtyVV(), GetPNoCom(), GradLogLForLambda(), Likelihood(), Load(), PrintSummary(), Save(), SeekJoin(), SeekLeave(), SeekSwitch(), SetDefaultPNoCom(), and SetPNoCom().
TFlt TAGMFit::RegCoef [private] |
Regularization parameter when we fit for P_c (for finding # communities).
Definition at line 22 of file agmfit.h.
Referenced by GradLogLForLambda(), Likelihood(), Load(), Save(), and SetRegCoef().
TRnd TAGMFit::Rnd [private] |
Definition at line 17 of file agmfit.h.
Referenced by Load(), RandomInit(), RandomInitCmtyVV(), RunMCMC(), and SampleTransition().